Abstract
TSD276-2, a wheat genetic stock derived from the cross Agra Local/T. spelta 276 showed broad spectrum resistance against leaf rust pathogen. Genetic analysis was undertaken using F1, F2, F2:3 and BC1F1 generations derived from the cross TSD276-2/Agra Local. The results revealed a single recessive gene for leaf rust resistance, tentatively named as LrTs276-2, in TSD276-2. Molecular mapping of leaf rust resistance gene LrTs276-2 in TSD276-2 was done using SNP-based PCR and SSR markers. For Bulked Segregant Analysis (BSA), two bulks viz. resistant bulk and susceptible bulk, and the parents TSD276-2 and Agra Local were genotyped for SNPs using AFFYMETRIX 35K Wheat Breeders' AXIOM array. T. spelta 276 was also genotyped and used as a check. BSA indicated that the gene for leaf rust resistance in TSD276-2 is located on chromosome arm 1DS. Putatively linked SNPs on chromosome arm 1DS were converted into PCR-based markers. Polymorphic SSR markers on chromosome arm 1DS were also identified. Final linkage map was constructed using one SNP-based PCR and three SSR markers. The rust reaction and chromosomal location suggest that LrTs276-2 is a new leaf rust resistance gene which may be useful in broadening the genetic base of leaf rust resistance in wheat.
Similar content being viewed by others
Introduction
Leaf rust caused by Puccinia triticina Eriks. is one of the most important and widespread foliar diseases of wheat (Triticum aestivum L.) inflicting significant yield losses in susceptible cultivars1,2,3,4. Although, rust diseases can be controlled by application of fungicides, genetic resistance remains the most effective, economical and environmentally sustainable method5,6. To date, 79 leaf rust resistance genes have been designated and catalogued in wheat and about half of them have their origin in various closely or distantly related species and genera of wheat while remaining resistance genes are native to wheat7,8,9. Many of the leaf rust resistance have become ineffective due to evolution of new virulent pathotypes. This necessitates continuous search for new and effective resistance genes for deployment in wheat cultivars. Spelt wheat (T. spelta) is potentially a good source of rust resistance genes10. T. spelta is a hulled wheat and is considered as ancestral to the free-threshing forms of hexaploid wheat11. Spelt wheat shares the same genomic structure, 2n = 6x = 42 (BBAADD genome) with common wheat and belongs to primary gene pool of wheat. This facilitates gene transfer from T. spelta by direct hybridization through homologous recombination. Till date, only three leaf rust resistance genes viz., Lr44 on chromosome 1B10, Lr65 on chromosome 2A12 and Lr71 on chromosome 1B13 from spelt wheat have been identified and mapped.
As part of our pre-breeding programme at Indian Agricultural Research Institute, New Delhi, we have been working to identify and map rust resistance genes from primary, secondary and tertiary gene pools14,15,16,17,18,19. The present study reports the inheritance and molecular mapping of leaf rust resistance in T. spelta derived bread wheat line TSD276-2.
Material and methods
Plant materials
Triticum spelta derived bread wheat line TSD276-2 and leaf rust susceptible cultivar Agra Local (AL) were used to study the mode of inheritance and molecular mapping of leaf rust resistance. TSD276-2 is derived from the cross T. spelta accession 276/Agra Local. T. spelta accession 276 (T. spelta276) is a winter wheat requiring either vernalization or a prolonged photoperiod for flowering while AL is spring wheat. TSD276-2 showed spring wheat nature with no vernalization requirement. The F1, F2 and F2:3 population from the cross TSD276-2/AL were used for genetic analysis and molecular mapping of leaf rust resistance. T. spelta 276, the donor of leaf rust resistance to TSD276-2 was also used as a check in this study.
Leaf rust pathotypes
Pure inoculum of Puccinia triticina pathotypes was obtained from ICAR-Indian Institute of Wheat and Barley Research, Regional Station, Flowerdale, Shimla. Pathotypes were multiplied and maintained on susceptible cultivar AL under greenhouse conditions at Division of Genetics, IARI, New Delhi. T. spelta 276 and its derivative TSD276-2 along with susceptible check Agra Local were tested with 17 diverse Puccinia triticina pathotypes during crop season 2017–2018 (Table 1). Pathotype 77-5 (121R63-1), currently one of the most predominant one in India, was used for genetic analysis and molecular mapping.
Screening for leaf rust resistance
Screening for leaf rust resistance was done at seedling stage in greenhouse. Seeds were sown in small rectangular metallic trays (28 cm × 10 cm × 7.5 cm). About 10 day old seedlings were inoculated by spraying an aqueous suspension of P. triticina uredospores. The uredospore suspension was mixed with a drop of Tween20. Inoculated seedlings were incubated in a humid glass chamber for 48 h and were subsequently transferred to benches in a greenhouse under ambient condition of light and relative humidity. Disease reaction was recorded 12 days after inoculation as per the method described by Stakman et al.20.
Molecular marker analysis
Fresh leaf samples collected from 40 to 45 days old plants were crushed in liquid nitrogen with mortar and pestle. DNA isolation was done following CTAB method21. DNA was quantified on 0.8% (w/v) agarose gel using Lambda Uncut DNA (THERMO FISHER SCIENTIFIC INC., USA) as standard and confirmed with NanoDrop Lite spectrophotometer (THERMO FISHER SCIENTIFIC INC., USA). DNA was diluted to the working stock concentration of 25 ng/μL and stored at − 20 °C.
For bulked segregant analysis equal amount of DNA from 20 homozygous resistant (HR) and 20 homozygous susceptible (HS) lines from F2:3 population was bulked to constitute two contrasting bulks viz. resistant bulk and susceptible bulk22. T. spelta276, TSD276-2, Agra Local and two extreme bulks i.e. RB and SB were genotyped for SNP using AFFYMETRIX 35K Wheat Breeders' AXIOM array23. SNPs found to be polymorphic between parents as well as bulks were identified. Chromosomal region found to show maximum polymorphic SNPs between bulks was presumed to carry leaf rust resistance gene. These SNPs were converted to PCR based markers using the software primer 3(v.0.4.0) as described earlier19. These SNP-based PCR markers were used for parental polymorphism.
Besides, a total 51 SSR markers spanning across the putatively identified chromosome 1D (on the basis of SNP genotyping) carrying rust resistance gene were tested for polymorphism between TSD276-2 and AL24,25. Primer sequences of these markers are available in public domain (https://wheat.pw.usda.gov/cgi-bin/GG3/browse.cgi?class=marker). For studying marker polymorphism between parents, PCR was performed in a reaction volume of 10 μl. Each 10 μl reaction volume included 2 μL of template DNA (50 ng), 1 μL forward primer (5 pm/μl), 1 μL of reverse primer (5 pm/μl), and 3 μL of Taq DNA Polymerase RED 2× master mix (AMPLIQON A/S, Denmark) and 3 μL of nuclease free water (THERMO FISHER SCIENTIFIC INC., USA). The PCR reactions were performed in 96-well PCR plates with thermal seal in an APPLIED BIOSYSTEMS VERITI thermal cycler at specific profile. The PCR conditions for primers used are given in Table 2. PCR amplified products were resolved on 3.5% (w/v) Agarose (LONZA, Rockland, USA) gel stained with ethidium bromide. Gels were visualized with a UV-transilluminator gel documentation system (SYNGENE G-BOX, Cambridge, UK).
Finally, both polymorphic SNP-based PCR markers and polymorphic SSR markers were used for bulked segregant analysis to confirm the identity of chromosome carrying leaf rust resistance gene22. Total 136 F2:3 homozygous lines were genotyped with SNP-based PCR marker as well as SSR markers identified as polymorphic in BSA. Linkage analysis was performed using MAPMAKER v3.026 with a minimum LOD score of 3.0 and a maximum genetic distance of 37.2 cM. ‘COMPARE’, ‘TRY’ and ‘RIPPLE’ commands of MAPMAKER v3.0 were used to check the final order of map. The genetic distances (cM) were calculated using the Kosambi mapping function27. Chi-square test was conducted to test the goodness-of-fit for segregation of the resistance gene28. Putative gene(s) present between flanking marker interval were predicted using wheat sequence (International Wheat Genome Sequencing Consortium, 2018) available at Ensembl Plants (https://plants.ensembl.org/Triticumaestivum/Info/Index) between the two flanking markers utilizing BioMart (https://plants.ensembl.org/biomart/martview/12b2b93c60bfbcedcaf0e4d1e023fee9).
Results
Genetic analysis of leaf rust resistance
TSD276-2 showed high degree of leaf rust resistance with ITs ranging from “0;” to “1−” against different P. triticina pathotypes, whereas the susceptible parent Agra Local showed susceptible reaction with infection type (IT) “3” to “33+” against all the pathotypes used in the study (Fig. 1). The original spelt wheat accession T. spelta 276 also showed high degree of resistance against all the 17 pathotypes (Table 1).
For genetic analysis, TSD276-2, AL, F1 (TSD276-2/AL) and 294 F2 plants were screened for leaf rust resistance against P. triticina pathotype 77-5. TSD276-2 showed resistance reaction with ITs “;1−N” whereas Agra Local showed susceptibility (IT 33+). All the 15 F1 plants were susceptible indicating recessive nature of resistance. Out of 294 F2 plants, 75 plants were resistant with ITs ranging from “;” to “1++” while 219 plants showed susceptible reaction (Fig. 1). The F2 segregation showed a good fit to theoretically expected ratio of 1 resistant: 3 susceptible plants (χ2(1:3) = 0.041, p-value = 0.84) for a single recessive gene. The results were further confirmed in F2:3 families. The 284 F2:3 families segregated into 1 resistant: 2 segregating: 1 susceptible with χ2(1:2:1) = 0.254 (p-value = 0.88). BC1 generation also showed expected segregation of IR:1S plants (Table 3).
Mapping of leaf rust resistance
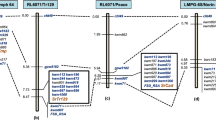
Genotyping data points for 35,143 SNP markers were obtained, which were filtered in a sequential manner. SNPs lacking any chromosome ID and position were removed. Moreover, SNPs showing heterozygous alleles are also filtered out. Further filtering resulted into 2414 SNPs showing polymorphism between parent viz. TSD276-2 and AL. Of these, only 20 SNPs were polymorphic between resistant and susceptible bulks. The 20 polymorphic SNPs were distributed over 10 chromosomes but five SNPs were observed in the short arm of chromosome 1D indicating putative linkage of these SNPs with leaf rust resistance gene in TSD276-2. The five SNPs on chromosome arm 1DS carried the identical alleles in T. spelta 276 and TSD276-2 (Table 4) and were converted into SNP-based PCR markers (Table 5). Among SSR markers on chromosome 1D, twelve were polymorphic between parents TSD276-2 and Agra Local. A combined BSA analysis using five SNP-based PCR markers and twelve SSR markers identified one SNP-based PCR marker (AX-94393474) and three SSR markers (Xcfd15, Xcfd61 and Xgwm106) as polymorphic between resistant and susceptible bulks (Fig. 2). For construction of linkage map, 136 F2:3 families comprising 68 homozygous resistant and 68 homozygous susceptible families were genotyped. Linkage map of leaf rust resistance gene in TSD276-2 was constructed with three SSR and one SNP-based PCR marker covering genetic distance of 18.7 cM on short arm of chromosome 1D (Fig. 3). The leaf rust resistance gene in TSD276-2, hereafter referred as LrTs276-2 is flanked by SSR markers Xcfd15 and Xcfd61 spanning 7.8 cM interval on map. SSR marker Xcfd15 mapped closest to the gene at 2.3 cM. The SNP based PCR marker AX-94393474 mapped 7.2 cM distal to the gene LrTs276-2. Xcfd61 mapped 5.5 cM proximal to rust resistance gene. The order of SSR and SNP based marker in the linkage map is consistent with the consensus map of Somers et al. 2004 as well as with CS-IWGSC RefSeq v1 (Table 5).
The SSR marker Xcfd15, closest to the resistance gene LrTs276-2 behaved as a codominant marker amplifying alleles of 180 and 200 bp in TSD276-2 and only one allele i.e. 168 bp in AL (Table 6, Supplementary Fig. S1 online). The 200 bp allele of SSR marker Xcfd15 was found to be linked with the leaf rust resistance in TSD276-2. T. spelta 276, the original source of resistance gene LrTs276-2 also amplified alleles identical to TSD276-2. The markers Xcfd61 and Xgwm106 were linked with leaf rust resistance gene in repulsion phase and behaved as dominant markers amplifying alleles of 195 bp and 127 bp in susceptible parent AL, respectively (Fig. 2). Both Xcfd61 and Xgwm106 produced null allele in TSD276-2 and T. spelta 276. The SNP-based PCR marker AX-94393474 was linked with resistance gene in coupling phase and amplified alleles of 211 and 232 bp in TSD276-2 and T. spelta 276 while only single allele i.e. 211 bp was produced in AL showing the dominant nature of marker (see Supplementary Fig. S1 online). Further, the total number of genes between two flanking markers i.e. cfd15 and cfd61 observed in Triticum aestivum (covering 6.36 Mb sequence) and Ae. tauschii (covering 6.21 Mb sequence) were 141 and 84, respectively. Out of these, 45 genes in Triticum aestivum and 27 genes in Ae. tauschii have R gene related domain (see Supplementary Table S1 and Supplementary Table S2 online). Among these, 24 genes are common between Triticum aestivum and Ae. tauschii between two flanking markers (see Supplementary Table S3 online).
Discussion
The leaf rust resistance gene in TSD276-2 showed a wide spectrum of resistance against Indian P. triticina pathotypes. Genetic analysis showed a single recessive gene conferring leaf rust resistance in TSD276-2. The resistance gene was mapped on short arm of chromosome 1D and was flanked by SSR markers Xcfd15 and Xcfd61. The resistance gene in TSD276-2 is derived from the spelt wheat accession T. spelta276. A large number of rust resistance genes have been transferred into wheat from the species belonging to secondary and tertiary gene pools. These resistance genes often carry some degree of linkage drag and sometimes undesirable genes17,29,30. Genetic resources from primary gene pool have the advantage of homologous recombination which can be used to remove the linkage drag. Closely related species of wheat from primary gene pool are rich and diverse source of unique alleles that can be used in wheat improvement12,31,32,33,34,35. In the present study, we identified a seedling leaf rust resistance gene tentatively named as LrTs276-2 in spelt wheat derived common wheat line TSD276-2. Till date, only three leaf rust resistance genes from spelt wheat viz., Lr44, Lr65 and Lr71 have been identified and mapped. While Lr44 and Lr71 have been located on chromosome 1B10,13, Lr65 has been mapped on chromosome 2A12. The leaf rust resistance gene in the present study has been mapped on short arm of chromosome 1D indicating that LrTs276-2 is different from the already characterized leaf rust resistance genes from spelt wheat and potentially a novel leaf rust resistance gene. Moreover, Lr44, Lr65 and Lr71 behaved as dominant genes while the LrTs276-2 in TSD276-2 is recessive in nature, differentiating this gene from other spelt wheat genes characterized so far. Lr65 has also been reported to be susceptible to Indian pathotype 77-519.
Till date, three leaf rust resistance genes viz., Lr2136,37, Lr4236,38,39,40 and Lr6041 have been mapped on 1DS chromosome of wheat. Among these genes Lr21 and Lr42 have been transferred in common wheat from diploid progenitor species Aegilops tauschii (2n = 2x = 14, genome DD)36,37,40,42 while Lr60 is native in bread wheat41,43. Although the gene LrTs276-2 mapped by us is from T. spelta and is expected to be different, nevertheless it is important to distinguish this gene from other leaf rust resistance genes mapped on chromosome 1DS. This can be done on the basis of differential response to P. triticina pathotypes and by comparing the genetic and physical position on the chromosome. The gene Lr21 is ineffective against several Indian pathotypes of P. triticina44 whereas in our study both T. spelta 276 and TSD276-2 showed high degree of resistance against all the 17 pathotypes used in the study. The response of Lr60 to Indian pathotypes of P. triticina is not available. However, Lr42 shows resistance to all the P. triticina pathotypes in India44. Hiebert et al.41 analyzed the linkage between Lr21 and Lr60 and observed that Lr60 is about 13.5 cM distal to Lr21 with the SSR marker Xbarc149 co-segregating with Lr21. Lr42 has been reported as race-specific partially dominant resistance gene36. However, Czembor et al.45 mapped Lr42 on chromosome 3D and observed that Lr42 behaved as dominant gene. Sun et al.38 mapped Lr42 on the distal end of chromosome arm 1DS and marker Xwmc432 was found closest to the gene Lr42 at a distance of 0.8 cM. Liu et al.39 reported Lr42 as recessive gene and mapped it on 1DS chromosome with flanking markers Xwmc432 and Xgdm33 spanning a genetic distance of 17 cM. Xwmc432 was the closest marker 4 cM proximal to Lr42. Gill et al.40 narrowed down the Lr42 region to 3.7 cM with marker TC387992 at a distance of 1.7 cM distal to Lr42 and Xwmc432 located at 2 cM proximal to Lr42. The gene LrTs276-2 mapped by us in TSD276-2 is flanked by the markers Xcfd15 and Xcfd61. The gene LrTs276-2 is 2.3 cM proximal to marker Xcfd15 while Gill et al.40 reported Lr42 at a distance of 5.4 cM distal to Xcfd15. Thus, Lr42 is located distal to LrTs276-2 on chromosome arm 1DS. A comparison of genetic and physical maps unambiguously shows that the locus represented by LrTs276-2 is different from other rust resistance loci mapped on chromosome 1DS (Fig. 4). Anchoring of markers linked to Lr21, Lr 42, Lr60 and LrTs276-2 on Chinese Spring Reference genome indicates that these genes are located on 1DS chromosome in order of Lr60-Lr21-Lr42 and LrTs276-2. Further, in-silico studies suggested that the flanking markers are more than 6 Mb apart. The predicted number of genes related to disease resistance with R gene domain (using domain reported by Peng et al.46) in the species i.e. Triticum aestivum and Aegilops tauschii are very high. Hence, it is essential to narrow down the region for prediction of putative candidate gene. The rust reaction, nature of gene and comparative genomics indicates that LrTs276-2 is a novel leaf rust resistance gene that may be useful in resistance breeding programs in wheat.
References
Marasas, C. N., Smale, M. & Singh, R. P. The economic impact in developing countries of leaf rust resistance breeding in CIMMYT-related spring bread wheat (CIMMYT, Mexico, DF, 2004).
Bolton, M. D., Kolmer, J. A. & Garvin, D. F. Wheat leaf rust caused by Puccinia triticina. Mol. Plant Pathol. 9, 563–575 (2008).
Huerta-Espino, J. et al. Global status of wheat leaf rust caused by Puccinia triticina. Euphytica 179, 143–160 (2011).
Tomar, S. M. S., Singh, S. K., Sivasamy, M. & Vinod. Wheat rusts in India, resistance breeding and gene deployment—A review. Indian J. Genet. 74, 129–156 (2014).
Kolmer, J. A. Genetics of resistance to wheat leaf rust. Annu. Rev. Phytopathol. 34, 435–455 (1996).
Keller, B. et al. Molecular analysis of fungal disease resistance in wheat. In Proceedings of the 11th International Wheat Genetics Symposium (eds Appels, R. et al.) (Sydney University Press Lynne McIntyre and Peter Sharp, Sydney, 2008).
Naik, B. K. et al. Molecular mapping and validation of the microsatellite markers linked to the Secale cereal derived leaf rust resistance gene Lr45 in wheat. Mol Breed. 35, 61 (2015).
McIntosh, R. A., Dubcovsky, J., Rogers, W. J., Morris, C. & Xia, X. C. Catalogue of Gene Symbols for Wheat 2017 Supplement (Komugi Wheat Genetic Resources Database, Yokohama, 2017).
Qureshi, N. et al. A new leaf rust resistance gene Lr79 mapped in chromosome 3BL from the durum wheat landrace Aus26582. Theor. Appl. Genet. 131, 1091–1098 (2018).
Dyck, P. L. & Sykes, E. E. Genetics of leaf-rust resistance in three spelt wheats. Can. J. Plant Sci. 74, 231–233 (1994).
McFadden, E. S. & Sears, E. R. The origin of Triticum spelta and its free-threshing hexaploid relatives. J. Hered. 37, 81–89 (1946).
Mohler, V., Singh, D., Singrun, C. & Park, R. F. Characterization and mapping of Lr65 in spelt wheat ‘Altgold Rotkorn’. Plant Breed. 131, 252–257 (2012).
Singh, D., Mohler, V. & Park, R. F. Discovery, characterisation and mapping of wheat leaf rust resistance gene Lr71. Euphytica 190, 131–136 (2013).
Gireesh, C., Vinod, Sharma, J. B. & Prabhu, K. V. Inheritance and molecular mapping of leaf rust resistance in Triticum turgidum var. durum cv. Trinakria. Indian J. Genet. 74, 10–15 (2014).
Gireesh, C., Vinod, Sharma, J. B. & Prabhu, K. V. Genetics and molecular mapping of stem rust resistance in bread wheat line WR95. Euphytica 205, 869–875 (2015).
Singh, A. K. et al. Genetics and mapping of a new leaf rust resistance gene in Triticum aestivum L. × Triticum timopheevii Zhuk. derivative ‘Selection G12’. J. Genet. 96, 291–297 (2017).
Niranjana, M. et al. Cytogenetic analysis and mapping of leaf rust resistance in Aegilops speltoides Tausch derived bread wheat line Selection 2427 carrying putative gametocidal gene(s). Genome 60, 1076–1085 (2017).
Nataraj, V. et al. Molecular characterization of Triticum militinae derived introgression lines carrying leaf rust resistance. Genet. Resour. Crop Evol. 65, 787–796 (2018).
Rani, K. et al. A novel leaf rust resistance gene introgressed from Aegilops markgrafii maps on chromosome arm 2AS of wheat. Theor. Appl. Genet. https://doi.org/10.1007/s00122-020-03625-w (2020).
Stakman, E. C., Stewart, D. M. & Loegering, W. Q. Identification of physiologic races of Puccinia graminis var. tritici. (Washington, USDA, 1962).
Murray, M. G. & Thompson, W. F. Rapid isolation of high molecular weight DNA. Nucleic Acids Res. 8, 4321–4325 (1980).
Michelmore, R. W., Paran, I. & Kesseli, R. V. Identification of markers linked to disease resistance genes by bulked segregant analysis, a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. 88, 9828–9832 (1991).
Allen, A. M. et al. Characterization of a Wheat Breeders’ Array suitable for high throughput SNP genotyping of global accessions of hexaploid bread wheat (Triticum aestivium). Plant Biotechnol. J. 15, 390–401 (2017).
Somers, D., Isaac, P. & Edwards, K. A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 109, 1105–1114 (2004).
Sourdille, P. et al. Microsatellite-based deletion bin system for the establishment of genetic-physical map relationships in wheat (Triticum aestivum L.). Funct. Integr. Genomics 4, 12–25 (2004).
Lander, E. S. et al. MAPMAKER, an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1, 174–181 (1987).
Kosambi, D. D. The estimation of map distances from recombination values. Ann. Eugen. 12, 172–175 (1943).
Mather, K. Measurement of Linkage in Heredity (Methuen and Company Limited, London, 1951).
Chhuneja, P. et al. Transfer of leaf rust and stripe rust resistance from Aegilops umbellulata Zhuk. to bread wheat (Triticum aestivum L.). Genet. Resour. Crop Evol. 55, 849–859 (2007).
Bansal, M. et al. Aegilops umbellulata introgression carrying leaf rust and stripe rust resistance genes Lr76 and Yr70 located to 9.47-Mb region on 5DS telomeric end through a combination of chromosome sorting and sequencing. Theor. Appl. Genet. 133, 903–915 (2020).
Srivastava, J. P. & Damania, A. B. Wheat Genetic Resources, Meeting Diverse Needs 57–64 (Wiley, Chichester, 1990).
Huang, X. Q., Kempf, H., Ganal, M. W. & Roder, M. S. Advanced backcross QTL analysis in progenies derived from a cross between a German elite winter wheat variety and a synthetic wheat (Triticum aestivum L.). Theor. Appl. Genet. 109, 933–943 (2004).
Gill, B. S. et al. Wheat genetics resource center: The first 25 years. Adv. Agron. 89, 73–136 (2006).
Gill, B. S. et al. Alien genetic resources for wheat leaf rust resistance, cytogenetic transfer, and molecular analysis. Aus. J. Agric. Res. 59, 197–205 (2008).
Ogbonnaya, F. C. et al. Synthetic hexaploids, harnessing species of the primary gene pool for wheat improvement. Plant Breed. Rev. 37, 35–122 (2013).
Cox, T. S., Raupp, W. J. & Gill, B. S. Leaf rust-resistance genes Lr41, Lr42, and Lr43 transferred from Triticum tauschii to common wheat. Crop Sci. 34, 339–343 (1994).
Huang, L. & Gill, B. S. An RGA-like marker detects all known Lr21 leaf rust resistance gene family members in Aegilops tauschii and wheat. Theor. Appl. Genet. 103, 1007–1013 (2001).
Sun, X., Bai, G., Carver, B. F. & Bowden, R. Molecular mapping of wheat leaf rust resistance gene Lr42. Crop Sci. 50, 59–66 (2010).
Liu, Z., Bowden, R. L. & Bai, G. Molecular markers for leaf rust resistance gene Lr42 in Wheat. Crop Sci. 53, 1566–1570 (2013).
Gill, H. S. et al. Fine mapping of the wheat leaf rust resistance gene Lr42. Int. J. Mol. Sci. 20, 2445 (2019).
Hiebert, C. W., Thomas, J. B., McCallum, B. D. & Somers, D. J. Genetic mapping of the wheat leaf rust resistance gene Lr60 (LrW2). Crop Sci. 48, 1020–1026 (2008).
Kerber, E. R. & Dyck, P. L. Inheritance in hexaploid wheat of leaf rust resistance and other characters derived from Aegilops squarrosa. Can. J. Genet. Cytol. 11, 639–647 (1969).
Dyck, P. L. Genetics of leaf rust resistance in 13 accessions of the Watkins wheat collection. Euphytica 80, 151–155 (1994).
Prasad, P. et al. Population differentiation of wheat leaf rust fungus Puccinia triticina in South Asia. Curr. Sci. 112(10), 2073–2084 (2017).
Czembor, P.C., Radecka-Janusik, M., Pietrusiñska, A. & Czembor, H. J. Mapping resistance gene to leaf rust in wheat line KSWGRC11 using Quantitative Bulked Segregant Analysis and DArT platform. In Proceedings of the 11th International Wheat Genetics Symposium (eds Appel, R. et al.) 739–740 (Sydney University Press, Sydney, 2008).
Peng, F. Y. & Yang, R. C. Prediction and analysis of three gene families related to leaf rust (Puccinia triticina) resistance in wheat (Triticum aestivum L.). BMC Plant Biol. 17, 108 (2017).
Acknowledgements
V.D. is grateful to UGC for financial assistance given as JRF and SRF during the course of study at ICAR-IARI, New Delhi and ICAR-World Bank funded National Agricultural Higher Education Project for providing financial assistance for SNP genotyping. The authors are grateful to the ICAR-Indian Institute of Wheat & Barley Research, Regional Station, Flowerdale, Shimla, for providing pure inoculum of P. triticina pathotypes and the ICAR-Indian Agricultural Research Institute, New Delhi, for facilitating the experiments.
Author information
Authors and Affiliations
Contributions
V.D. and S.K.J.—population development, rust screening, marker analysis, data acquisition, data analysis and interpretation, drafting of manuscript; N.M. and M.N.—drafting and review of manuscript; P.A.—marker analysis and drafting of the manuscript; J.B.S.—rust screening and multipathotype testing; V.—concept and design of study, study supervision, data interpretation, drafting and review of manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dinkar, V., Jha, S.K., Mallick, N. et al. Molecular mapping of a new recessive wheat leaf rust resistance gene originating from Triticum spelta. Sci Rep 10, 22113 (2020). https://doi.org/10.1038/s41598-020-78679-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-78679-3
This article is cited by
-
Characterization and identification of sources of rust resistance in Triticum militinae derivatives
Scientific Reports (2024)
-
Multi-locus genome-wide association studies (ML-GWAS) reveal novel genomic regions associated with seedling and adult plant stage leaf rust resistance in bread wheat (Triticum aestivum L.)
Heredity (2022)
-
Quick mapping and characterization of a co-located kernel length and thousand-kernel weight-related QTL in wheat
Theoretical and Applied Genetics (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.